Next Generation Sequencing 3: Purifying DNA Samples with Magnetic Beads - Eric Chow (UCSF)
https://www.ibiology.org/techniques/n...
Next generation sequencing allows DNA samples to be sequenced quickly and affordably. Learn how next gen sequencing works and get tips on preparing and running your samples.
In the past decade there has been an amazing change in the efficiency of DNA sequencing. Using traditional Sanger sequencing, the human genome project took 20 years and cost $3 billion. Current next generation sequencing methods allow a human genome to be sequenced for $1000, in 48 hours! In this talk, Eric Chow explains the chemistry behind next generation sequencing, and describes how the next gen sequencers detect and display results. The most commonly used Illumina sequencers are image based and detect the addition of fluorescently labelled nucleotides. Chow also describes two different next generation sequencing technologies which provide benefits such as much longer reads but with downsides such as higher error rates. Chow finishes the talk with some insights into medical applications of next gen sequencing such as much less invasive prenatal testing or cancer detection.
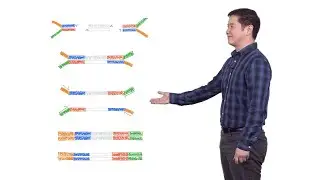
In two short how-to videos, Chow gives advice on purifying DNA samples using magnetic beads and on determining the quality of your nucleic acid sample using an Agilent Bioanalyzer.
Speaker Biography:
Eric Chow is an assistant professor in the Department of Biochemistry and Biophysics and the Director of the Center for Advanced Technology (CAT) at the University of California, San Francisco. The CAT provides resources for UCSF labs wishing to use next generation sequencing techniques and Chow’s research program strives to develop new applications for NGS in pathogen diagnostics. Chow received his BA in molecular biology from the University of California, Berkeley and his PhD in biochemistry from UCSF.